One of the more astonishing rhetorical gambits of objectors to the design inference is to try to suggest that the alphanumeric, code-using, algorithmic information system we see in the D/RNA of the living cell and linked protein synthesis is not really an information system, it all reduces to chemical reaction trains. A common associated gambit is to assert that terms like “code” etc are all readily dismissible analogies.
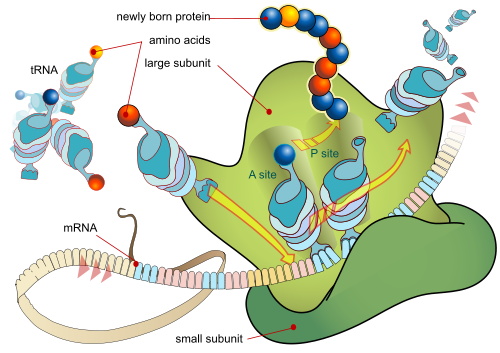
As a first reminder, protein synthesis as graphically summarised:
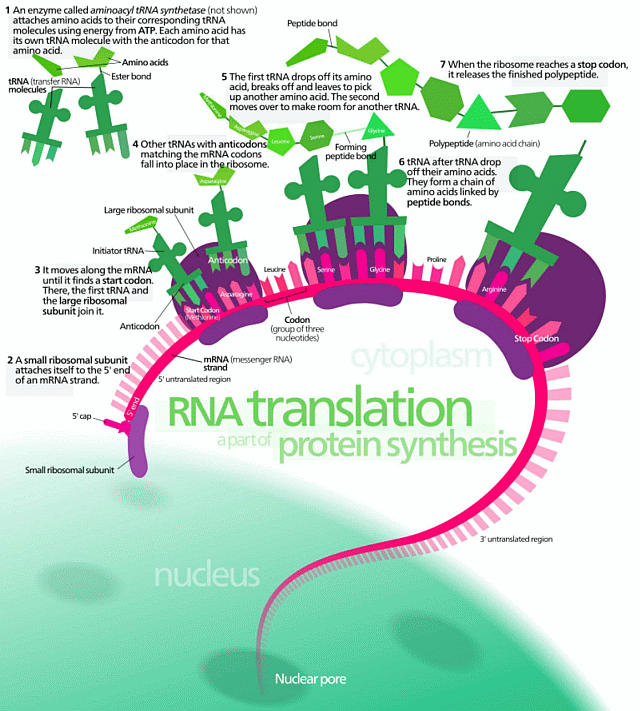
Of course, it never hurts to remind such objectors of p. 5 of Sir Francis Crick’s $6 million, March 19, 1953 letter to his son, Michael:

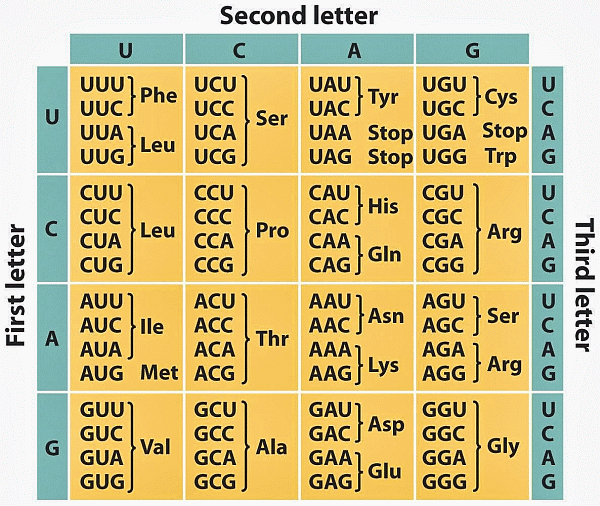
Notice, his belief right from the outset of discovering the double-helix stricture: “. . . D.N.A. is a code.” This was then drawn out by the early 1970’s, yielding several Nobel Prizes along the way, and it is now a commonplace from primary school on. Here is the “standard” code as tabulated in RNA form, one of about two dozen dialects:
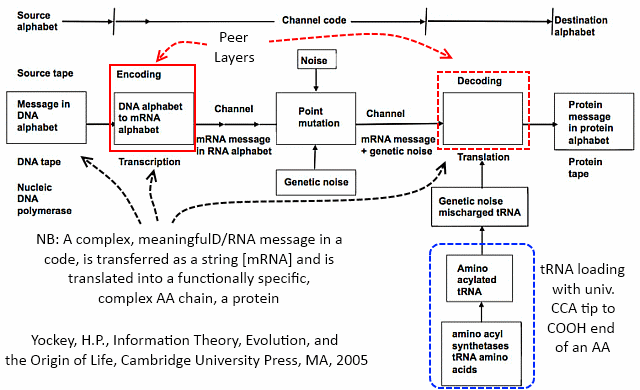
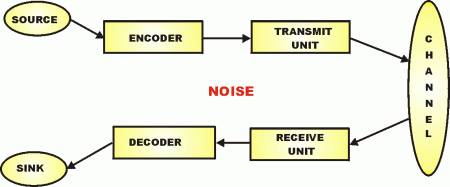
Here, too, is how Yockey fits the Protein synthesis framework into a standard communication system model:
That brings us to the role of tRNA, as we can see in the lower right, dotted blue box. Notice, the remark that tRNA is loaded with the AA required in protein synthesis, at a universal CCA tool-tip, which is at the 3′ end of the primary tRNA chain. That is, in effect, Chemically, any AA can be loaded at the tip of any tRNA. Indeed, there are cases — such as H pylori (the ulcer germ) — where an AA is loaded then chemically altered to the true target and experimenters have used the universality to add other AAs than the usual twenty found in nature.
Let us therefore look at the plan view, “clover-leaf” secondary structure of tRNA, noting how H-bonding sets up the emerging tool-arm structure:

Also, we can see how the CCA tip [at the A end] attaches to the COOH end of the AA, how the anti-codon is at the opposite end in one of the “loops,” and how it folds into its tertiary, 3-D position-arm structure:

Let me add, it is helpful to look at the structure of an amino acid, which allows modular chaining:

I find a series of videos helpful. First, charging/ loading the tRNA:
Next, translation — yes, a linguistic information-processing process carried out operationally using molecular nanotech (instead of the Si we use in microprocessors in the fetch, decode, execute loop):
Next, for completeness, the ribosome, first as a simplified diagram:
Video, with background details:
Fourthly, initiation of actual protein synthesis:
With all of this on the table, I can now headline a comment I made in the Ortho keyboard thread [a thread that is well worth working through], which — predictably — was side stepped rhetorically by objectors:
[KF, 29:] >>I’m late to the party. tRNA folds into a secondary cloverleaf structure then into the arm shape with the anticodon loop at one end and the standard CCA tool-tip that bonds to the loaded AA [COOH end] at the other. [break, let’s remind ourselves of the cloverleaf:]

As the tip is a standard [CCA–> COOH] joint, it is non-specific. That’s not the magic step.
That oh so humble source, Wikipedia, by principle of embarrassment, now tells an astonishing point or two of truth on how:
Aminoacylation is the process of adding an aminoacyl group to a compound. It covalently links an amino acid to the CCA 3′ end of a tRNA molecule. Each tRNA is aminoacylated (or charged) with a specific amino acid by an aminoacyl tRNA synthetase. There is normally a single aminoacyl tRNA synthetase for each amino acid, despite the fact that there can be more than one tRNA, and more than one anticodon for an amino acid. Recognition of the appropriate tRNA by the synthetases is not mediated solely by the anticodon, and the acceptor stem often plays a prominent role.[17] Reaction:
amino acid + ATP –> aminoacyl-AMP + PPi
aminoacyl-AMP + tRNA –> aminoacyl-tRNA + AMP
In some ways, even more tellingly, we find evidence of adaptation to apparent loss of function and/or alternative pathways:
Certain organisms can have one or more aminoacyl tRNA synthetases missing. This leads to charging of the tRNA by a chemically related amino acid, and by use of an enzyme or enzymes, the tRNA is modified to be correctly charged. For example, Helicobacter pylori has glutaminyl tRNA synthetase missing. Thus, glutamate tRNA synthetase charges tRNA-glutamine(tRNA-Gln) with glutamate. An amidotransferase then converts the acid side chain of the glutamate to the amide, forming the correctly charged gln-tRNA-Gln.
This stage is when the correlation between anticodon and targetted AA is assigned. Notice, it is not solely based on the anticodon, reflects conformation of the folded tRNA and in some cases has a workaround compensating for somehow missing information that leads to absence of a particular loading enzyme. H pylori loads a chemically related AA to the true target then brings up enzymes to modify to the correct AA. I have already noted that artificial AAs have been inserted and are capable of loading then being added to peptide chains.
It bears noting that in the tertiary structure, the anticodon and AA are at opposite ends of the tRNA. Which AA is there is set in a way that is modular and there can even be workarounds.
This is of course the point where a mobile, molecular scale position-arm device is loaded with the properly coded AA for protein synthesis. (I assume we can look that up for ourselves.)
Wiki’s disclosures against known ideological bent are not finished, we now go to the ribosome, the molecular machine that assembles a peptide chain based on the mRNA tape, using the coding.
Notice, the position-arm action at molecular level:
The ribosome has three binding sites for tRNA molecules that span the space between the two ribosomal subunits: the A (aminoacyl),[19] P (peptidyl), and E (exit) sites. In addition, the ribosome has two other sites for tRNA binding that are used during mRNA decoding or during the initiation of protein synthesis. These are the T site (named elongation factor Tu) and I site (initiation).[20][21] . . . .
Once translation initiation is complete, the first aminoacyl tRNA is located in the P/P site, ready for the elongation cycle described below. During translation elongation, tRNA first binds to the ribosome as part of a complex with elongation factor Tu (EF-Tu) or its eukaryotic (eEF-1) or archaeal counterpart. This initial tRNA binding site is called the A/T site. In the A/T site, the A-site half resides in the small ribosomal subunit where the mRNA decoding site is located. The mRNA decoding site is where the mRNA codon is read out during translation. The T-site half resides mainly on the large ribosomal subunit where EF-Tu or eEF-1 interacts with the ribosome. Once mRNA decoding is complete, the aminoacyl-tRNA is bound in the A/A site and is ready for the next peptide bond to be formed to its attached amino acid. The peptidyl-tRNA, which transfers the growing polypeptide to the aminoacyl-tRNA bound in the A/A site, is bound in the P/P site. Once the peptide bond is formed, the tRNA in the P/P site is deacylated, or has a free 3’ end, and the tRNA in the A/A site carries the growing polypeptide chain. To allow for the next elongation cycle, the tRNAs then move through hybrid A/P and P/E binding sites, before completing the cycle and residing in the P/P and E/E sites. Once the A/A and P/P tRNAs have moved to the P/P and E/E sites, the mRNA has also moved over by one codon and the A/T site is vacant, ready for the next round of mRNA decoding. The tRNA bound in the E/E site then leaves the ribosome.
The P/I site is actually the first to bind to aminoacyl tRNA, which is delivered by an initiation factor called IF2 in bacteria.[21] However, the existence of the P/I site in eukaryotic or archaeal ribosomes has not yet been confirmed. The P-site protein L27 has been determined by affinity labeling by E. Collatz and A.P. Czernilofsky (FEBS Lett., Vol. 63, pp. 283–86, 1976).
This is a tape-controlled assembly process dependent on the correctly loaded tRNAs to operate correctly. Where, proteins are the workhorse molecules of the cell.
Indeed, we effectively have a transfer machine with a miniature assembly line, with preloaded parts attached to position-arm carriers brought up and applied then exiting even as the target of the manufacturing process is being produced and held in place until complete.
Illustrating:

The mRNA tape, of course, carries initiation, stepwise elongation, finite succession and halting. Thus we see an algorithm at work, with an information bearing tape as controller and an assembly line using mobile position-arm units.
This is seriously advanced automation and manufacturing, using algorithms and code on string data structures [let’s only mention editing to form mRNAs], thus language. Where the coding is present twice in two separate subsystems, processed effectively independent of one another. DNA is unzipped and used to assemble mRNA precursors, which are edited to form the control tape. Separately, tRNA is created with the implicit anticodon, again stored in the master tape, DNA. Properly folded tRNAs are loaded with the appropriate AA or a precursor that is modified to be correct.
These are then brought into the ribosome under proper manufacturing control and peptide chains are built for further formation into proteins used in cellular processes including this one.
Chicken-egg loops abound, pointing to FSCO/I and islands of function requiring initial manufacture to a design, on pain of fruitlessly, aimlessly wandering in seas of non function and exhausting the blind search capability of the observed cosmos.
At this stage, it is manifest why a design inference on protein synthesis is robust and empirically grounded. As for means:
clever designer[s] + molecular nanotech lab –> clever FSCO/I rich design
Of course, one is free to reject such, but in all responsible fairness needs to provide a cogent, empirically warranted explanation.>>
Such an empirically warranted, blind watchmaker blind chance and/or mechanical necessity explanation remains conspicuously absent, even as agent explanations are ideologically locked out, never mind the significance of LANGUAGE and ALGORITHMS here.
Oh. yes, let me give a simplified overview of the context of protein synthesis:
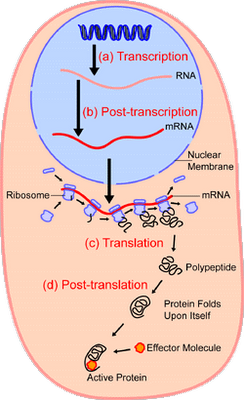
So, now, we can see the molecular nanotech in action and how it uses a base of Chemical-Physical processes, but we actually have a layer-cake architecture, once code enters as codes are based on arbitrary symbols that have to be decoded on the receiver side of a system. Echoing Yockey as seen above, the general architecture:
Thus, we see how we can stack codes and protocols:
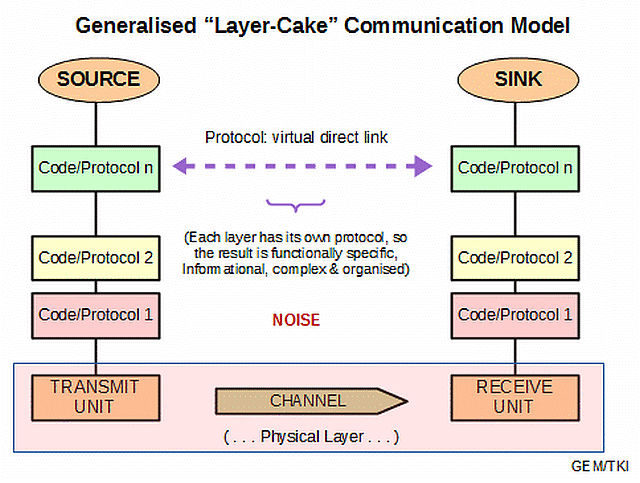
In terms of information-processing machines, this layer-cake stack becomes a stack of virtual machines, through which the design of the obvious, physical layer is developed to enable the virtual interactions:
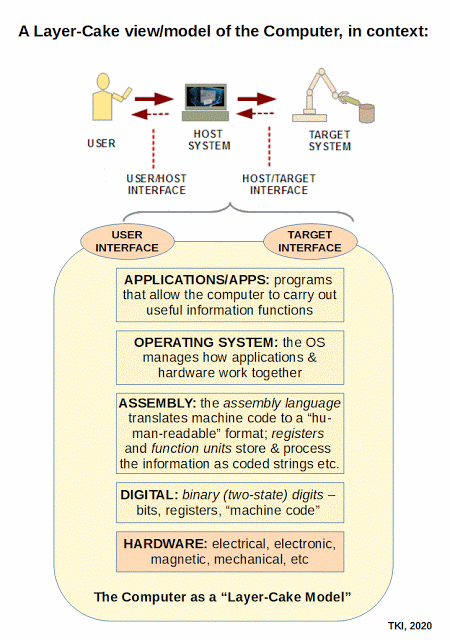
This brings out one of the key underlying fallacies of the dominant evolutionary materialistic scientism ideology: its mind-blindness that leads it to over-emphasise the physical layer, failing to see that that layer is shaped by needs, purposes and considerations relating to design. So, if such is imposed, it invites selectively hyperskeptical lockout of the implications of evidence that shows higher, information, meaning and purpose layers plausibly arising due to language-using agency in action. Where, language reduced to machine code is a manifestation of language at the physical level.
A further error is to then play the ideological, “gold standard” dismissible or even “no evidence” gambit. Having blinkered oneself ideologically, one then is selectively hyperskeptical and dismisses or denigrates inconvenient evidence. Then, the metaphysical question-begging is announced as “there’s no evidence that X . . .” — where, X takes on any inconvenient value.
Later in the thread, we see a case:
[EG, 57:] >>KF
It is noteworthy that algorithmic, alphanumeric code — a linguistic phenomenon — remains stubbornly as only the product of intelligence.
In fact, to the best of our knowledge, it remains stubbornly as only the product of human intelligence.>>
Obviously, the molecular nanotech in the living cell is antecedent to not only human life but cell based life. To this, I therefore responded:
[KF, 59:] >> you know full well that no one takes seriously the notion that we exhaust possibilities for intelligent beings. Looking to linked logic of being, we exemplify intelligence but in no way that implies exhausting it. Therefore the rhetorical gambit to try to suggest that cases of human intelligence forbid us from inferring characteristics of intelligence that could go beyond humans, is nonsense. That this has been used as a common atheistical talking point despite cogent correction only underscores the weakness of the atheistical case. Especially, when the issue on the table is linguistic, algorithmic, alphanumerical code not only antecedent to human life but constitutive of cell based life . . . >>
Attempted piling on:
[JVL, 60 :] >>KF: EG, you know full well that no one takes seriously the notion that we exhaust possibilities for intelligent beings.
But the only intelligent beings we know of that are even close to accomplishing highly sophisticated feats of engineering are human beings. We’ve got no living quarters, no spacecraft, no midden piles, no processing plants, pretty much nothing.
I’m perfectly happy to guess that there are other intelligent beings in the universe but, so far, we have zero evidence they exist. And we certainly have no evidence they have visited Earth.>>
My onward response:
[KF, 61:] >> we are contingent, intelligent, responsible and rational. We exemplify what is possible, we do not exhaust it. So, as you know or should know, it is an abuse of inductive reasoning to infer from our case or to suggest from our case that we exhaust possibilities for such beings. This becomes all the more blatantly fallacious and even desperate when it is used as a rhetorical gambit to try to blunt the evidence that in the heart of the living cell is alphanumeric, algorithmic, linguistic code. And that evidence alone — the evidence of the whole world of cell based life — is striking and has been striking since March 19, 1953. This is not an “isolated”readily dismissed case, we are talking about a central, keystone aspect of cell based life. The rhetorical resort is utterly telling on the force of the evidence and on the ruinous nature of selective hyperskepticism and resulting refusal to entertain the force of evidence. Precisely, what we have now seen live with the “gold standard” fallacy in the face of a pandemic and mounting evidence. So, this is not merely an academic oddity, it is a ruinous error we are dealing with. >>
EARLY U/D: While this OP was being developed, there was another comment:
[JVL, 62:] >>Kairosfocus: So, as you know or should know, it is an abuse of inductive reasoning to infer from our case or to suggest from our case that we exhaust possibilities for such beings.
I’m not, I’m just saying that we have no evidence of such beings except for humans.
The rhetorical resort is utterly telling on the force of the evidence and on the ruinous nature of selective hyperskepticism and resulting refusal to entertain the force of evidence.
Yes but you haven’t been completely successful getting most scientists to agree with your evidence. In the world common forum we’d have to call that disputed at best.>>
The evidence in question is the Genetic Code, as in, a standard term, there for a cause. Where, appeal to one sided “consensus” on a matter of controversy only underscores the need to actually face rather than evade evidence. My response:
[KF, 65:] >> JVL, the “no evidence” gambit is a signature selectively hyperskeptical fallacy. The evidence has been on the table, taught in every school, for coming on two full generations. As Crick noted from the outset, in the form of a belief: ” . . . the D.N.A. IS a code” and that belief was then drawn out in multiple Nobel Prize winning work over the next 20 years. The evidence you are trying to dismiss is the molecular nanotech, prong height [ . . . similar to von Neumann’s kinematic self replicator . . . ] 4-state per symbol code used algorithmically to build proteins, and requiring chicken and egg causal loop molecular nanomachines constituting a von Neuman kinematic self replicator. That is, despite denials and dismissals, LANGUAGE is at the heart of the design for cell based life. That has to be faced as we contemplate the protein synthesis machine code and its dozens of dialects. The repeated attempt to dismiss simply inadvertently underscores the force of the point and how telling it is. >>
Let’s add, an illustration of what a vNSR is like . . . and note, this was proposed in 1948, so for example the tape element and the assembly machinery [where tRNA is a position-arm assembly element that works in the ribosome as an assembly line transfer machine]:

Transfer machines? Oh, lesee again:

Refresher, Ribosome in action:

Oh, but I saved the best for the last. UB replies to EG on the same set of rhetorical stunts:
[UB, 64:] >>#51
Ed, sometimes you just try too hard. It’s a bad look.
Allow me to remind you of something that you seem to forget with incredible regularity. We are all human beings here, Ed. On average we were all pushed and prodded in our youth to develop a lifelong sense of judgement and to make proper distinctions between things like right and wrong, truth and fiction, vice and virtue, and so on, and so forth. It is one of the things that is most common between us, regardless of our cultures, and with any luck, by the time we reach maturity, we end up with a healthy guiding experience of being part of, and watching, human relationships. Among those many common experiences is the experience of someone who simply will not answer a valid question. This is an inevitable experience where not answering the question actually becomes their answer. We understand this predicament and we recognize what it looks like in our human brothers and sisters – with all the denials and the (often flagrant) excuses and the deflections of this and that and the other. What I am getting at here Ed, is that we all know what cookie crumbs in a little boy’s bed actually means, as a very simple example which any parent would understand.
Now Ed, you have been told that a physicist can practice their craft and measure the physical system that enables the use of language. A physicist can measure that system by its necessary physical entailments (and their relationships to one another). A physicist can therefore exclusively identify the use of language among other physical systems, and the gene system has been thus identified. This has been done. Furthermore, you’ve told that the physicists measurement and identification is a confirmation of a previous prediction – a prediction that the gene system would necessarily function by way of encoded language, which is itself an additional confirmation on the very nature of such systems. In other words, Ed, the conclusion that the gene system uses a code (for crying out loud) is a conclusion that comes through science without even a hair out of place.
Having to stoop to “Francis Crick was sloppy” as your next maneuver (to avoid the physics) is therefore quite a sight to see. It makes you look weak, poorly motivated, and as common as you can humanly be, just as non-answers often do. You should probably stop using that excuse.>>
Houston, we’ve got a problem. END
PS: I cannot but give a promotion to a comment by UD newbie (as a commenter . . . by his admission, he’s been lurking for years), Chris Messier, noting also the too often “misunderestimated” BA77’s contribution . . . as in, go read the thread. Chris:
[CM, 12:] >>First of all, thank you to BA77 for all the amazing info I’ve gleaned from your comments over the years. So much of the material you cite is _way_ over my head, but I take what I can manage
DaveS wrote:
“I’m severely underqualified for this discussion, but I wouldn’t mind seeing more detail on this”
I’m also very unqualified, so maybe I can give it a try, since I really like the whole DNA coding-mRNA-tRNA-protein pathway.
If I’m understanding what the author meant,
//There is a point in time and space where an association is made between a codon and an anticodon.//
That is, inside the ribosome the mRNA codon is ‘read’ by a tRNA molecule
with the matching anticodon. That’s the first ‘association’.
The next one
//There is also a point in time and space when there is an association made between an anticodon and an amino acid.//
Simply put, that is accomplished by the tRNA molecule. But to get tRNA
you need the aminoacyl tRNA synthetases, one for each of the 20 tRNA
molecules
An animation of protein synthesis
https://youtu.be/kmrUzDYAmEI
A short one on aminoacyl tRNA synthetases
https://youtu.be/180_sM9iYVk
Two related montages I made. The first one Joe DeWeese covers tRNA
https://youtu.be/sOI5u01LwyQ
The second one Stephen Meyer describes Francis Crick’s elucidating the need for translation of DNA in order to get to proteins, through what he called the ‘adaptor hypothesis’
https://youtu.be/rDLPjxzt1YE>>
PPS: LM, too, is well worth pondering at 11:
[LM, 11:] >>I sometimes want to throw up my hands in exasperation watching you all talk past each other. Let me approach it another way though I don’t expect success.
Let’s return to Sev’s original comment to the OP. (Edited for brevity.)
Seversky@1
Seriously?? You are both denying the material/physical structure and functions of the genome and a computer, that there is no physical chain of events between me pressing the ‘A’ key on my keyboard and the letter ‘A’ appearing on my screen? ….
Yes Sev, there is a nice neat physical chain of events from the ‘A’ key to the pattern of pixels that are displayed on my screen. Odd the pattern I see on the keyboard looks nothing like what shows up on the screen. It’s gone from an upside down ‘V’ with a bar across the middle to a small circle with a bar on the right. Encoded one way, output completely different. We’ll set that aside for just a bit.
Now let’s compare that to the cell. Again what’s happening is physical. There’s a series of DNA bases that when physically read by the right enzyme in the presence of enough of the right kind of amino acids will output a protein. There’s a vague similarity in your example to the transfer of the ‘A’ key (plastic with inlaid white marks) to its unlike ‘a’ (with light and dark pixels).
But wait! We’ve left something out. Who selected the ‘A’ key before pressing it? Why it was Sev. He’s an integral part of the whole process. And who recognized the output as having meaning? Of the thousands and thousands of possible shapes (fonts) that could represent the concept of ‘A’ who recognized ‘a’ as a representative of the concept of ‘A’? I believe that was Sev again!
Who selected those base pairs to represent that particular protein? Don’t you wonder about the Who that is just as much a part of the processes in the cell as the who was that picked that key?
Col. 1:17 He is before all things, and in him all things hold together.>>
Copyright © 2020 Uncommon Descent. This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.
Plugin by Taragana